Introduction
Apoptosis is a controlled and highly regulated form of cell death that is vital for normal embryonic development and for maintaining healthy cell populations. Assays that identify apoptosis form a critical and widely used toolkit to investigate the different stages of programmed cell death, to understand its regulation in normal development and its dysregulation in diseases. Dysregulated apoptosis is central in the development of cancer, tissue atrophy and several neurodegenerative diseases, providing an important therapeutic target in research.
Apoptosis mechanism
Apoptosis is initiated by internal or external cellular signals which ultimately instigate a common pathway leading to cellular and nuclear degradation via proteolytic caspases.
While a cell is undergoing apoptosis several morphological changes occur which can be visualised through microscopy. These include changes such as cell cytoplasm shrinkage, chromosome condensation (pyknosis), cell rounding and blebbing due to the breakdown of the cytoskeleton by caspases. There are also less obvious cellular changes occurring alongside, which include changes to the phospholipid membrane composition, loss of mitochondrial membrane potential and the activation of proteolytic caspases which degrade the cellular components (Figure 1). Thus there are numerous targets for the investigation of apoptosis in both healthy development and disease.
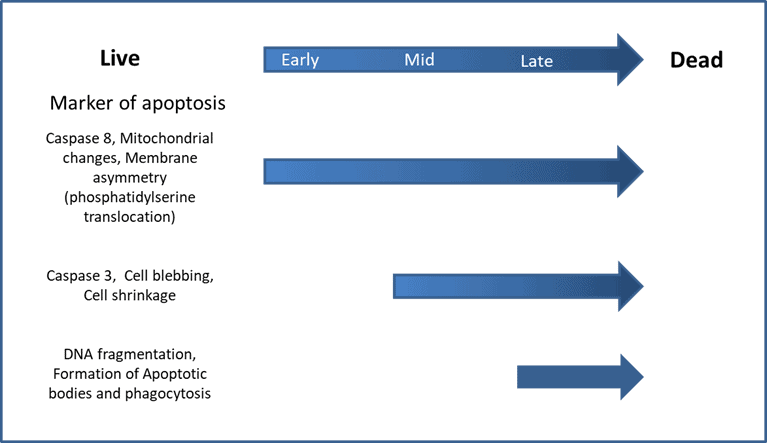 Figure 1. Early, mid and late stages of apoptosis. The early stages include the activation of Caspase 8, loss of mitochondrial membrane potential and movement of phosphatidylserine to the outer membrane leaflet. Mid stages include the activation of caspase 3 and cell shrinkage and blebbing. The end stages include the fragmentation of DNA, formation of apoptotic bodies and ultimately the phagocytosis by macrophages.
Figure 1. Early, mid and late stages of apoptosis. The early stages include the activation of Caspase 8, loss of mitochondrial membrane potential and movement of phosphatidylserine to the outer membrane leaflet. Mid stages include the activation of caspase 3 and cell shrinkage and blebbing. The end stages include the fragmentation of DNA, formation of apoptotic bodies and ultimately the phagocytosis by macrophages.
Apoptosis assays
Many common assays have been developed in the past few decades which rely on the use of fluorescent based imaging probes to identify markers of apoptosis using microscopy. These probes often fluoresce in the presence of apoptotic markers, such as activated caspases or phosphatidylserine (PS) in the outer leaflet of the membrane. For in vitro experiments, these studies often include a step where cells are treated with compounds which may induce or inhibit apoptosis. The cells can then be imaged with a fluorescence microscope and quantified using image analysis software.
The different types of assays can be broadly split into groups based on their mechanism, such as mitochondrial function, membrane asymmetry, caspase activation or DNA changes.
Mitochondrial function assays. Mitochondrial disruption occurs early in the cell death process. Reagents that accumulate in healthy (active) mitochondria such as membrane potential dependant probes (e.g. rhodamine 123, tetra-methylrhodamine) can be used to assess mitochondrial health in live cells (Lu, 2018). Mitochondrial disruption includes loss of membrane potential through the mitochondrial permeability transition pore (MPTP), which can be measured using JC-1 fluorescent dyes (Sivandzade, 2019). Loss of membrane potential causes changes to mitochondrial calcium levels and oxidative stress, which can be detected by fluorescent calcium indicators, such as Rhod-2 AM, and superoxide detectors (mitosoxtm) (Dikalov, 2014; Davidson, 2018). Cytochrome c is also released into the cytoplasm during mitochondrial disruption and can be detected by immunofluorescence staining (Matapurkar, 2006).
Membrane asymmetry and phospholipid translocation. Once apoptosis is initiated PS translocates from the inner face of the plasma membrane to the outer leaflet providing a marker for apoptosis. Annexin V binds with high affinity to negatively charged phospholipids (i.e. PS) so fluorescently tagged Annexin V (e.g. Annexin V-FITC) can identify this translocation of PS during apoptosis (Zhang, 2018).
Caspase activation. The activation of caspases is a critical step in the apoptosis pathway. Certain probes exist which bind to activated caspases, these are known as Fluorescence Inhibitor of Caspase Activity (FLICA), such as those found in ImageIT Live Caspase detection kits. Other probes introduce a caspase substrate that produces fluorescence upon cleavage by active caspases, such as CellEvent Caspase-3/7 (Lee, 2019; Darzynkiewicz Z, 2011).
Chromatin condensation and DNA fragmentation assays. Chromatin condensation can be visualized with a generic nuclear stain such as DAPI or Hoechst (Crowley, 2016). DNA fragmentation can be visualized using in-situ staining of DNA strand breaks in the terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL) assay, using a nucleotide tagged with a fluorescent dye (Loo, 2011).
Specific knockout assays. Specific components of the apoptosis pathway can be knocked out, using CRISPR or transient siRNA inhibition. This removes the activity of a protein to understand how it affects the apoptotic process. It was through knock out studies that caspase 3 was identified as a key mediator of apoptosis in mammals (Zheng, 2013).
Applications of apoptosis assays
The applications of apoptosis assays are extremely varied ranging from investigative research, disease pathogenesis, diagnostics and drug discovery.
Therapeutically, interest lies in both causing apoptosis, to remove unwanted unhealthy cells, and preventing unintentional apoptosis, to prevent the death of healthy cells.
Apoptosis inhibition causes cells to ignore internal or external stimuli that would normally initiate apoptosis. This leads to the survival and proliferation of damaged cells which increases the likelihood of diseases like cancer. Therefore, in drug discovery, many anti-cancer strategies focus on the restoration of apoptosis to cause cell death in these specific cell populations and shrink tumours.
Conversely, unwanted cell death in healthy tissues is a hallmark of many disease processes including tissue atrophy, HIV infection and neurodegenerative conditions like Alzheimer’s and Parkinson’s disease. Research into these areas has highlighted the role apoptosis plays in disease pathogenesis, providing a significant target for new, more effective therapies.
Apoptosis assays can also be employed as a control for ensuring cell health during drug discovery, or in fact, any routine cell study. This makes them widely applicable in all areas of microscopical research. Any cell-based experiment that involves imaging changes to a cell population can benefit from the use of a simple cell death, or cell viability, marker to ensure the experimental conditions do not cause unintentional apoptosis.
High Content Imaging
High content imaging and high content screening are employed to increase throughput for apoptosis assays. This provides a powerful tool in research to examine many compounds in parallel using 96 or 384 well plates to those that may cause cell death, either intentionally or unintentionally, or prevent it. This can be further combined with automated image analysis to segment cell populations of interest (i.e. those undergoing particular stages of apoptosis) and extract information about their fluorescence intensity or localisation, size and shape. This can be automatically quantified during acquisition using computational software to give fast, accurate results with minimal user input.
Summary
Apoptosis is highly controlled cell death that is executed by cellular caspases following internal or external stimuli. It is critical for normal development, but is also often dysregulated in a variety of diseases, providing a crucial target for therapy.
Assays that identify apoptosis are vital to a host of fields, from personalized cancer therapies to determining disease mechanisms. Techniques such as fluorescent labelling and microscopy are critical for imaging the apoptotic process, providing necessary insight into the complex interplay of pro and anti-apoptotic factors.
Advancements such as the automation of imaging apoptosis assays in parallel, combined with real-time computational analysis, are providing new opportunities, revolutionizing this field of research.
References
Crowley, L. M. (2016). Analyzing Cell Death by Nuclear Staining with Hoechst 33342. Cold Spring Harbor Protocols, 6(9).
Darzynkiewicz Z, P. P. (2011). Fluorochrome-labeled inhibitors of caspases: convenient in vitro and in vivo markers of apoptotic cells for cytometric analysis. Methods in Molecular Biology.
Davidson, S. a. (2018). Imaging Mitochondrial Calcium Fluxes with Fluorescent Probes and Single- or Two-Photon Confocal Microscopy. Methods in Molecular Biology, 171-186.
Dikalov, E. I. (2014). Methods for Detection of Mitochondrial and Cellular Reactive Oxygen Species. Antioxidants and Redox Signalling, 20(2), 372-382.
Ferlini, C. a. (2007). Assay for apoptosis using the mitochondrial probes, Rhodamine123 and 10-N-nonyl acridine orange. Nature Protocols, 2(12), 3111-3114.
Lee, S. Y. (2019). A High Throughput Apoptosis Assay using 3D Cultured Cells. Molecules, 24(18).
Loo, D. T. (2011). In Situ Detection of Apoptosis by the TUNEL Assay: An Overview of Techniques. Methods in Molecular Biology (Methods and Protocols), 682.
Lu, J. W. (2018). Detection of Mitochondria Membrane Potential to Study CLIC4 Knockdown-induced HN4 Cell Apoptosis In Vitro. Journal of Visualized Experiments(137).
Matapurkar, A. a. (2006). Requirement of Cytochrome C for Apoptosis in Human Cells. Cell Death and Differentiation, 13, 2062-2067.
McKenzie, M. L. (2018). Simultaneous Measurement of Mitochondrial Calcium and Mitochondrial Membrane Potential in Live Cells by Fluorescent Microscopy. Journal of Visualized Experiments(119).
Sivandzade, F. B. (2019). Analysis of the Mitochondrial Membrane Potential Using the Cationic JC-1 Dye as a Sensitive Fluorescent Probe. Bio-protocol, 9(1), e3128.
Yang, L. M.-h. (2003). Predominant Suppression of Apoptosome by Inhibitor of Apoptosis Protein in Non-Small Cell Lung Cancer H460 Cells. Cancer Research, 63, 831-837.
Zhang, G. G. (2018). Early Detection of Apoptosis Using a Fluorescent Conjugate of Annexin V. Biotechniques, 23(3), 525-531.
Zheng, T. S. (2013). Learning from Deficiency: Gene Targeting of Caspases. Madame Curie Bioscience Database. Austin (Texas): Landes Bioscience.